publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2026
-
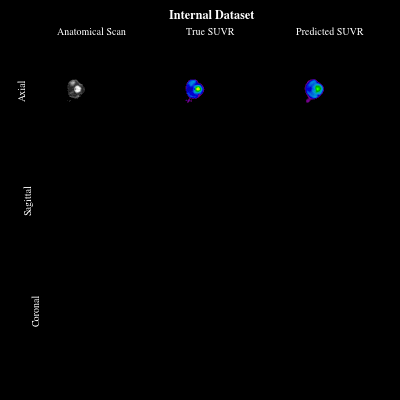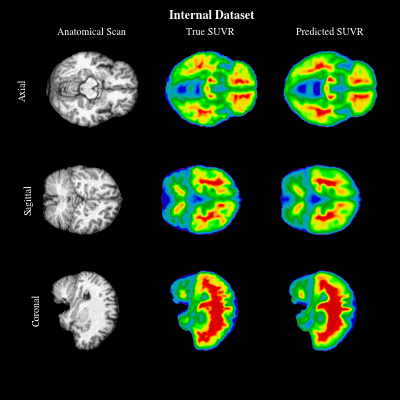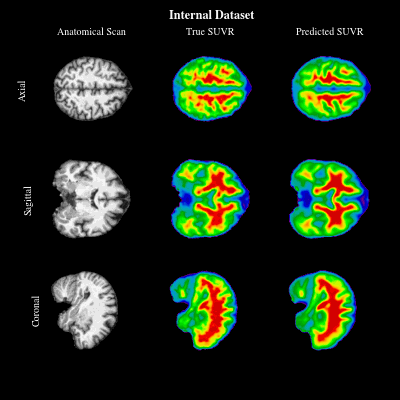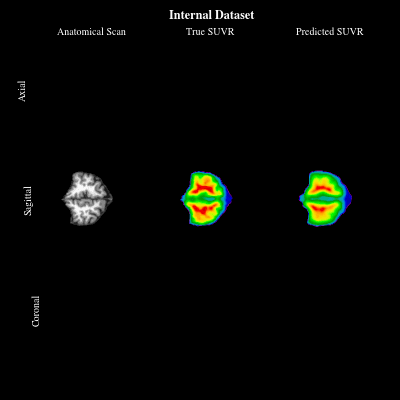 CoCoLIT: ControlNet-Conditioned Latent Image Translation for MRI to Amyloid PET SynthesisAlec Sargood†, Lemuel Puglisi†, James H Cole, and 3 more authorsIn AAAI, 2026† Joint first authorship (<17% acceptance rate over 20k+ papers)
CoCoLIT: ControlNet-Conditioned Latent Image Translation for MRI to Amyloid PET SynthesisAlec Sargood†, Lemuel Puglisi†, James H Cole, and 3 more authorsIn AAAI, 2026† Joint first authorship (<17% acceptance rate over 20k+ papers)Synthesizing amyloid PET scans from the more widely available and accessible structural MRI modality offers a promising, cost-effective approach for large-scale Alzheimer’s Disease (AD) screening. This is motivated by evidence that, while MRI does not directly detect amyloid pathology, it may nonetheless encode information correlated with amyloid deposition that can be uncovered through advanced modeling. However, the high dimensionality and structural complexity of 3D neuroimaging data pose significant challenges for existing MRI-to-PET translation methods. Modeling the cross-modality relationship in a lower-dimensional latent space can simplify the learning task and enable more effective translation. As such, we present CoCoLIT (ControlNet-Conditioned Latent Image Translation), a diffusion-based latent generative framework that incorporates three main innovations: (1) a novel Weighted Image Space Loss (WISL) that improves latent representation learning and synthesis quality; (2) a theoretical and empirical analysis of Latent Average Stabilization (LAS), an existing technique used in similar generative models to enhance inference consistency; and (3) the introduction of ControlNet-based conditioning for MRI-to-PET translation. We evaluate CoCoLIT’s performance on publicly available datasets and find that our model significantly outperforms state-of-the-art methods on both image-based and amyloid-related metrics. Notably, in amyloid-positivity classification, CoCoLIT outperforms the second-best method with improvements of +10.5% on the internal dataset and +23.7% on the external dataset.
-
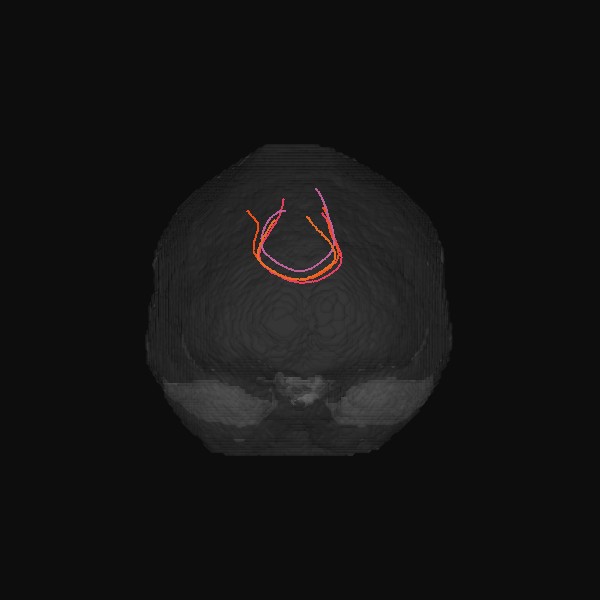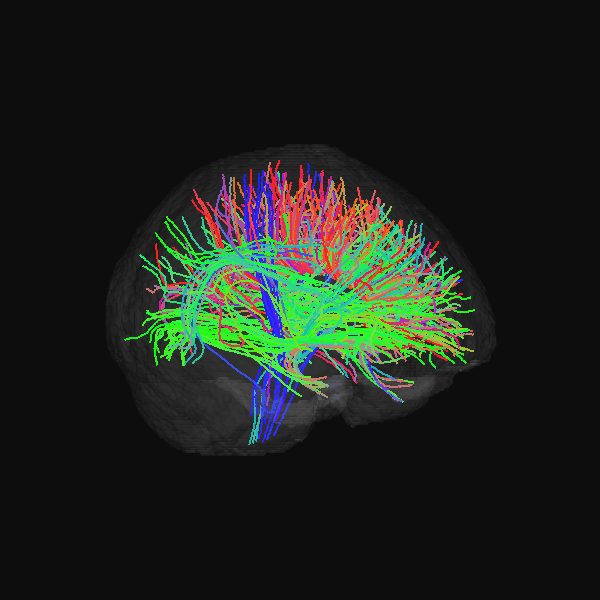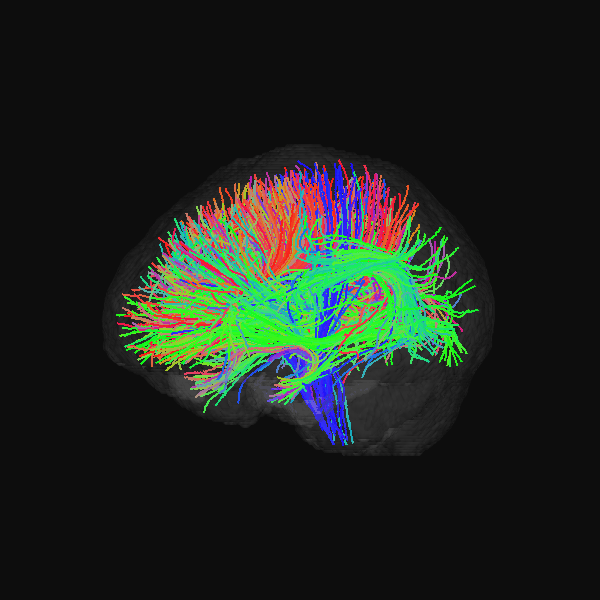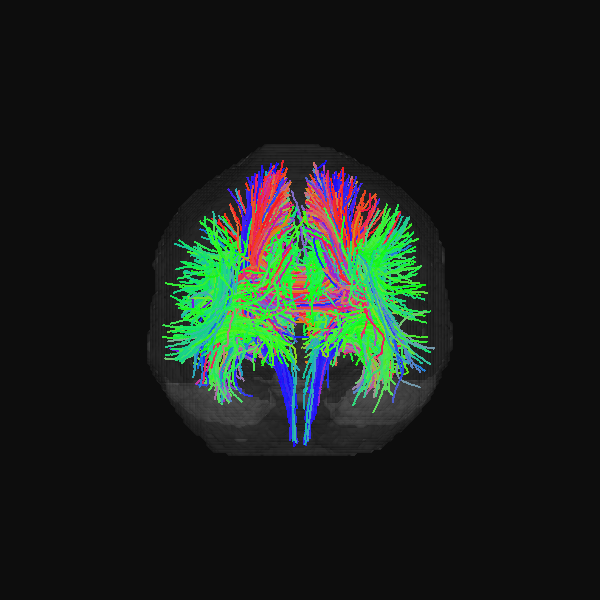 GenTract: Generative Global TractographyAlec Sargood, Lemuel Puglisi, Elinor Thompson, and 2 more authorsIn Arxiv Preprint, 2026Alec Sargood’s promising work on global tractography.
GenTract: Generative Global TractographyAlec Sargood, Lemuel Puglisi, Elinor Thompson, and 2 more authorsIn Arxiv Preprint, 2026Alec Sargood’s promising work on global tractography.Tractography is the process of inferring the trajectories of white-matter pathways in the brain from diffusion magnetic resonance imaging (dMRI). Local tractography methods, which construct streamlines by following local fiber orientation estimates stepwise through an image, are prone to error accumulation and high false positive rates, particularly on noisy or low-resolution data. In contrast, global methods, which attempt to optimize a collection of streamlines to maximize compatibility with underlying fiber orientation estimates, are computationally expensive. To address these challenges, we introduce GenTract, the first generative model for global tractography. We frame tractography as a generative task, learning a direct mapping from dMRI to complete, anatomically plausible streamlines. We compare both diffusion-based and flow matching paradigms and evaluate GenTract’s performance against state-of-the-art baselines. Notably, GenTract achieves precision 2.1x higher than the next-best method, TractOracle. This advantage becomes even more pronounced in challenging low-resolution and noisy settings, where it outperforms the closest competitor by an order of magnitude. By producing tractograms with high precision on research-grade data while also maintaining reliability on imperfect, lower-resolution data, GenTract represents a promising solution for global tractography.
-
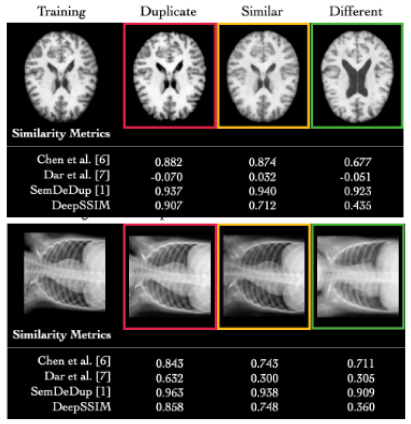 A Novel Metric for Detecting Memorization in Generative Models for Brain MRI SynthesisAntonio Scardace†, Lemuel Puglisi†, Francesco Guarnera, and 2 more authorsIn WACV, 2026† Joint first authorship
A Novel Metric for Detecting Memorization in Generative Models for Brain MRI SynthesisAntonio Scardace†, Lemuel Puglisi†, Francesco Guarnera, and 2 more authorsIn WACV, 2026† Joint first authorshipDeep generative models have emerged as a transformative tool in medical imaging, offering substantial potential for synthetic data generation. However, recent empirical studies highlight a critical vulnerability: these models can memorize sensitive training data, posing significant risks of unauthorized patient information disclosure. Detecting memorization in generative models remains particularly challenging, necessitating scalable methods capable of identifying training data leakage across large sets of generated samples. In this work, we propose DeepSSIM, a novel self-supervised metric for quantifying memorization in generative models. DeepSSIM is trained to: i) project images into a learned embedding space and ii) force the cosine similarity between embeddings to match the ground-truth SSIM (Structural Similarity Index) scores computed in the image space. To capture domain-specific anatomical features, training incorporates structure-preserving augmentations, allowing DeepSSIM to estimate similarity reliably without requiring precise spatial alignment. We evaluate DeepSSIM in a case study involving synthetic brain MRI data generated by a Latent Diffusion Model (LDM) trained under memorization-prone conditions, using 2,195 MRI scans from two publicly available datasets (IXI and CoRR). Compared to state-of-the-art memorization metrics, DeepSSIM achieves superior performance, improving F1 scores by an average of +52.03% over the best existing method. Code and data of our approach are publicly available at the following link: https://github.com/brAIn-science/DeepSSIM.
2025
-
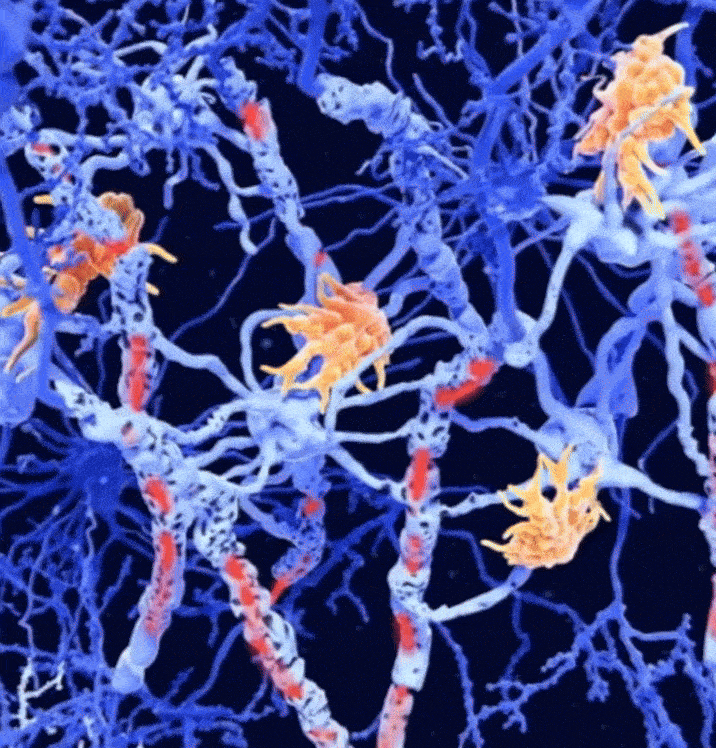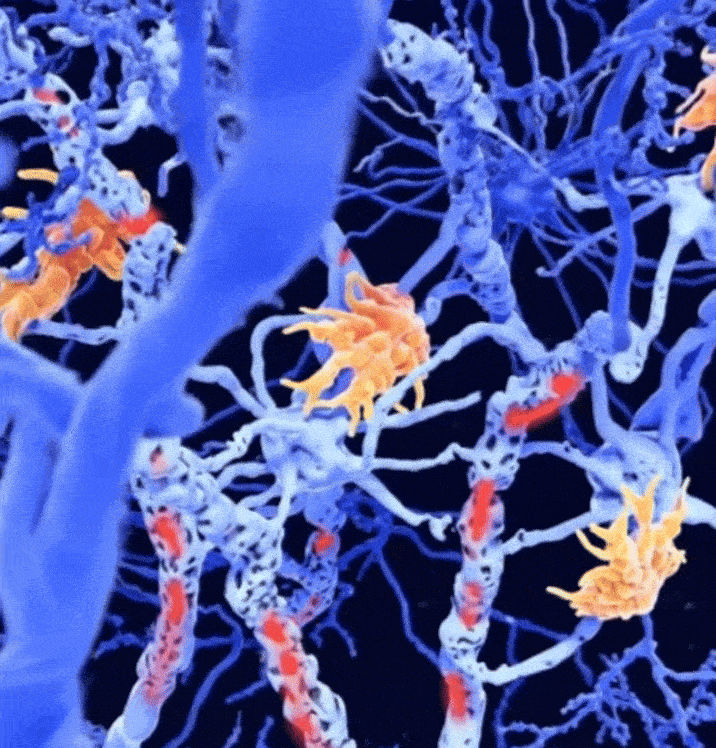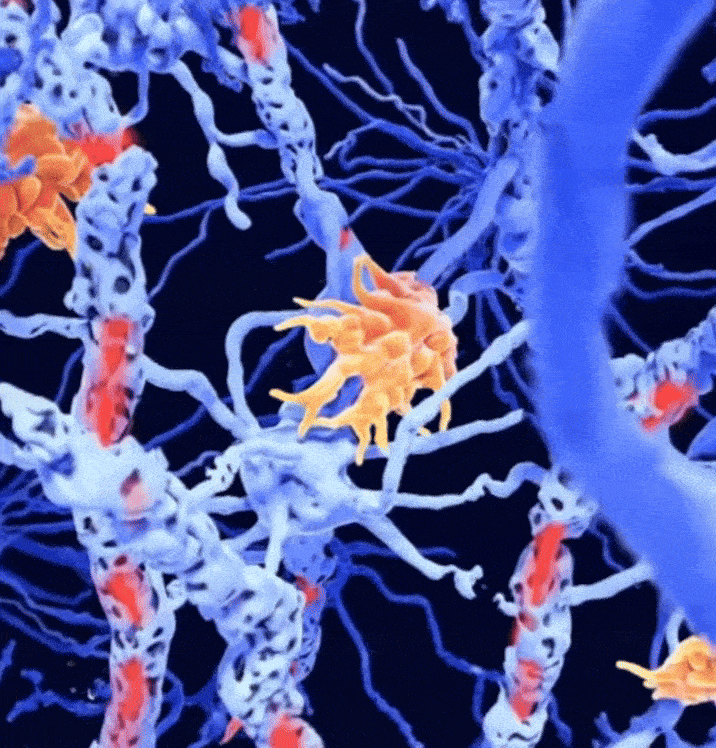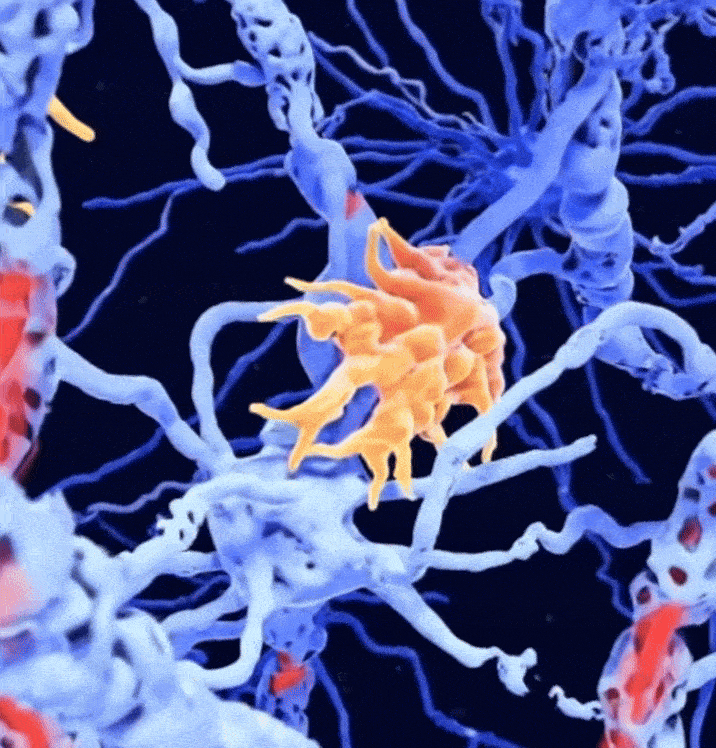 Combined magnetic resonance imaging and serum analysis reveals distinct multiple sclerosis typesCharles Willard†, Lemuel Puglisi†, Daniele Ravi, and 7 more authorsIn Brain, 2025† Joint first authorship; 🌟 Featured in The Guardian.
Combined magnetic resonance imaging and serum analysis reveals distinct multiple sclerosis typesCharles Willard†, Lemuel Puglisi†, Daniele Ravi, and 7 more authorsIn Brain, 2025† Joint first authorship; 🌟 Featured in The Guardian.Multiple sclerosis (MS) is a highly heterogeneous disease in its clinical manifestation and progression. Predicting individual disease courses is key for aligning treatments with underlying pathobiology. We developed an unsupervised machine learning model integrating MRI-derived measures with serum neurofilament light chain (sNfL) levels to identify biologically informed MS subtypes and stages. Using a training cohort of patients with relapsing–remitting and secondary progressive MS (n = 189), with validation on a newly diagnosed population (n = 445), we discovered two distinct subtypes defined by the timing of sNfL elevation and MRI abnormalities (early- and late-sNfL types). In comparison to MRI-only models, incorporating sNfL with MRI improved correlations of data-derived stages with the Expanded Disability Status Scale in the training (Spearman’s ρ = 0.420 versus MRI-only ρ = 0.231, P = 0.001) and external test sets (ρ = 0.163 for MRI–sNfL, versus ρ = 0.067 for MRI-only). The early-sNfL subtype showed elevated sNfL, corpus callosum injury and early lesion accrual, reflecting more active inflammation and neurodegeneration, whereas the late-sNfL group showed early volume loss in the cortical and deep grey matter volumes, with later sNfL elevation. Cross-sectional subtyping predicted longitudinal radiological activity: the early-sNfL group showed a 144% increased risk of new lesion formation (hazard ratio = 2.44, 95% confidence interval 1.38–4.30, P < 0.005) compared with the late-sNfL group. Baseline subtyping, over time, predicted treatment effect on new lesion formation on the external test set (faster lesion accrual in early-sNfL compared with late-sNfL, P = 0.01), in addition to treatment effects on brain atrophy (early sNfL average percentage brain volume change: −0.41, late-sNfL = −0.31, P = 0.04). Integration of sNfL provides an improved framework in comparison to MRI-only subtyping of MS to stage disease progression and inform prognosis. Our model predicted treatment responsiveness in early, more active disease states. This approach offers a powerful alternative to conventional clinical phenotypes and supports future efforts to refine prognostication and guide personalized therapy in MS.
-
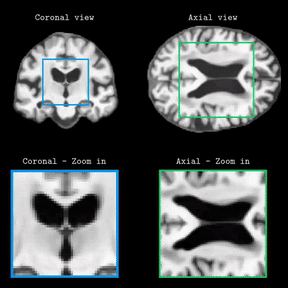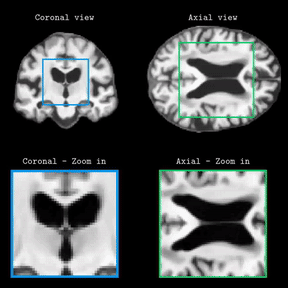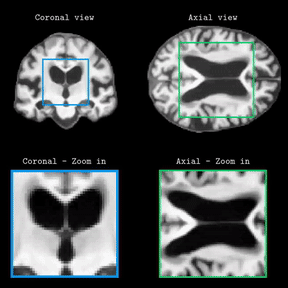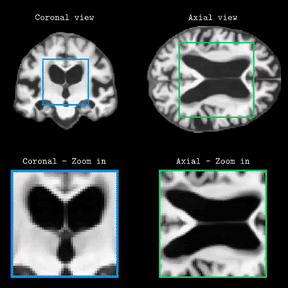 Brain Latent Progression: Individual-based Spatiotemporal Disease Progression on 3D Brain MRIs via Latent DiffusionLemuel Puglisi, Daniel C Alexander, and Daniele RavìIn Medical Image Analysis, 2025Impact Factor = 11.8; 🌟 Runner-up @ Best MedIA Paper Award
Brain Latent Progression: Individual-based Spatiotemporal Disease Progression on 3D Brain MRIs via Latent DiffusionLemuel Puglisi, Daniel C Alexander, and Daniele RavìIn Medical Image Analysis, 2025Impact Factor = 11.8; 🌟 Runner-up @ Best MedIA Paper AwardThe growing availability of longitudinal Magnetic Resonance Imaging (MRI) datasets has facilitated Artificial Intelligence (AI)-driven modeling of disease progression, making it possible to predict future medical scans for individual patients. However, despite significant advancements in AI, current methods continue to face challenges including achieving patient-specific individualization, ensuring spatiotemporal consistency, efficiently utilizing longitudinal data, and managing the substantial memory demands of 3D scans. To address these challenges, we propose Brain Latent Progression (BrLP), a novel spatiotemporal model designed to predict individual-level disease progression in 3D brain MRIs. The key contributions in BrLP are fourfold: (i) it operates in a small latent space, mitigating the computational challenges posed by high-dimensional imaging data; (ii) it explicitly integrates subject metadata to enhance the individualization of predictions; (iii) it incorporates prior knowledge of disease dynamics through an auxiliary model, facilitating the integration of longitudinal data; and (iv) it introduces the Latent Average Stabilization (LAS) algorithm, which (a) enforces spatiotemporal consistency in the predicted progression at inference time and (b) allows us to derive a measure of the uncertainty for the prediction at the global and voxel level. We train and evaluate BrLP on 11,730 T1-weighted (T1w) brain MRIs from 2,805 subjects and validate its generalizability on an external test set comprising 2,257 MRIs from 962 subjects. Our experiments compare BrLP-generated MRI scans with real follow-up MRIs, demonstrating state-of-the-art accuracy compared to existing methods. The code is publicly available at: https://github.com/LemuelPuglisi/BrLP.
2024
-
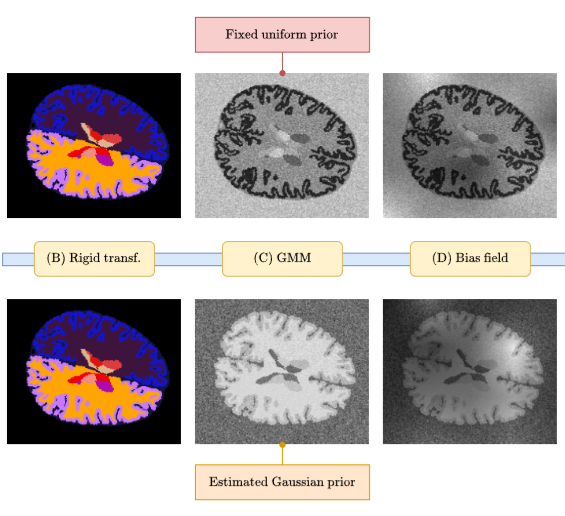 SynthBA: Reliable Brain Age Estimation Across Multiple MRI Sequences and ResolutionsLemuel Puglisi, Alessia Rondinella, Linda De Meo, and 3 more authorsIn IEEE Metrology for eXtended Reality, Artificial Intelligence and Neural Engineering, 2024
SynthBA: Reliable Brain Age Estimation Across Multiple MRI Sequences and ResolutionsLemuel Puglisi, Alessia Rondinella, Linda De Meo, and 3 more authorsIn IEEE Metrology for eXtended Reality, Artificial Intelligence and Neural Engineering, 2024Brain age is a critical measure that reflects the biological ageing process of the brain. The gap between brain age and chronological age, referred to as brain PAD (Predicted Age Difference), has been utilized to investigate neurodegenerative conditions. Brain age can be predicted using MRIs and machine learning techniques. However, existing methods are often sensitive to acquisition-related variabilities, such as differences in acquisition protocols, scanners, MRI sequences, and resolutions, significantly limiting their application in highly heterogeneous clinical settings. In this study, we introduce Synthetic Brain Age (SynthBA), a robust deep-learning model designed for predicting brain age. SynthBA utilizes an advanced domain randomization technique, ensuring effective operation across a wide array of acquisition-related variabilities. To assess the effectiveness and robustness of SynthBA, we evaluate its predictive capabilities on internal and external datasets, encompassing various MRI sequences and resolutions, and compare it with state-of-the-art techniques. Additionally, we calculate the brain PAD in a large cohort of subjects with Alzheimer’s Disease (AD), demonstrating a significant correlation with AD-related measures of cognitive dysfunction. SynthBA holds the potential to facilitate the broader adoption of brain age prediction in clinical settings, where re-training or fine-tuning is often unfeasible. The SynthBA source code and pre-trained models are publicly available at https://github.com/LemuelPuglisi/SynthBA.
-
 Enhancing Spatiotemporal Disease Progression Models via Latent Diffusion and Prior KnowledgeLemuel Puglisi, Daniel C Alexander, and Daniele RavìIn International Conference on Medical Image Computing and Computer Assisted Intervention, 2024🌟 Oral, Best Paper Award Candidate (top <1%)
Enhancing Spatiotemporal Disease Progression Models via Latent Diffusion and Prior KnowledgeLemuel Puglisi, Daniel C Alexander, and Daniele RavìIn International Conference on Medical Image Computing and Computer Assisted Intervention, 2024🌟 Oral, Best Paper Award Candidate (top <1%)In this work, we introduce Brain Latent Progression (BrLP), a novel spatiotemporal disease progression model based on latent diffusion. BrLP is designed to predict the evolution of diseases at the individual level on 3D brain MRIs. Existing deep generative models developed for this task are primarily data-driven and face challenges in learning disease progressions. BrLP addresses these challenges by incorporating prior knowledge from disease models to enhance the accuracy of predictions. To implement this, we propose to integrate an auxiliary model that infers volumetric changes in various brain regions. Additionally, we introduce Latent Average Stabilization (LAS), a novel technique to improve spatiotemporal consistency of the predicted progression. BrLP is trained and evaluated on a large dataset comprising 11,730 T1-weighted brain MRIs from 2,805 subjects, collected from three publicly available, longitudinal Alzheimer’s Disease (AD) studies. In our experiments, we compare the MRI scans generated by BrLP with the actual follow-up MRIs available from the subjects, in both cross-sectional and longitudinal settings. BrLP demonstrates significant improvements over existing methods, with an increase of 22% in volumetric accuracy across AD-related brain regions and 43% in image similarity to the ground-truth scans. The ability of BrLP to generate conditioned 3D scans at the subject level, along with the novelty of integrating prior knowledge to enhance accuracy, represents a significant advancement in disease progression modeling, opening new avenues for precision medicine. The code of BrLP is available at the following link: https://github.com/LemuelPuglisi/BrLP.
-
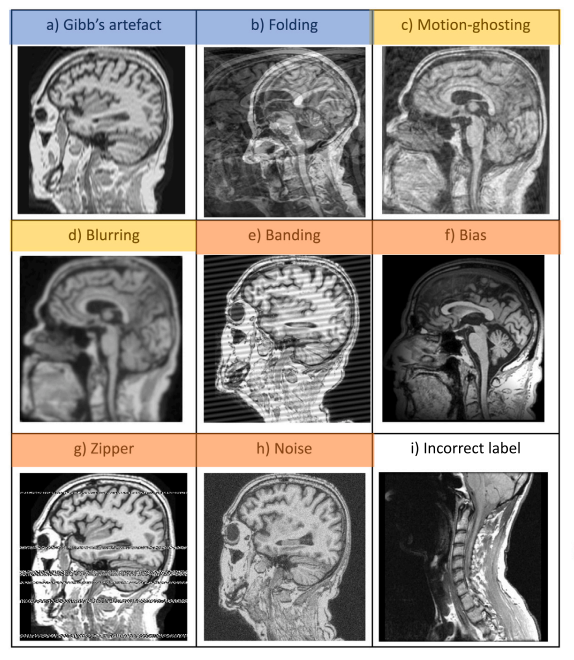 An efficient semi-supervised quality control system trained using physics-based MRI-artefact generators and adversarial trainingDaniele Ravi, Frederik Barkhof, Daniel C. Alexander, and 3 more authorsIn Medical Image Analysis, 2024Impact Factor = 11.8
An efficient semi-supervised quality control system trained using physics-based MRI-artefact generators and adversarial trainingDaniele Ravi, Frederik Barkhof, Daniel C. Alexander, and 3 more authorsIn Medical Image Analysis, 2024Impact Factor = 11.8Large medical imaging data sets are becoming increasingly available, but ensuring sample quality without significant artefacts is challenging. Existing methods for identifying imperfections in medical imaging rely on data-intensive approaches, compounded by a scarcity of artefact-rich scans for training machine learning models in clinical research. To tackle this problem, we propose a framework with four main components: 1) artefact generators inspired by magnetic resonance physics to corrupt brain MRI scans and augment a training dataset, 2) abstract and engineered features to represent images compactly, 3) a feature selection process depending on the artefact class to improve classification, and 4) SVM classifiers to identify artefacts. Our contributions are threefold: first, physics-based artefact generators produce synthetic brain MRI scans with controlled artefacts for data augmentation. This will avoid the labour-intensive collection and labelling process of scans with rare artefacts. Second, we propose a pool of abstract and engineered image features to identify 9 different artefacts for structural MRI. Finally, we use an artefact-based feature selection block that, for each class of artefacts, finds the set of features providing the best classification performance. We performed validation experiments on a large data set of scans with artificially-generated artefacts, and in a multiple sclerosis clinical trial where real artefacts were identified by experts, showing that the proposed pipeline outperforms traditional methods. In particular, our data augmentation increases performance by up to 12.5 percentage points on accuracy, precision, and recall. The computational efficiency of our pipeline enables potential real-time deployment, promising high-throughput clinical applications through automated image-processing pipelines driven by quality control systems.
2023
-
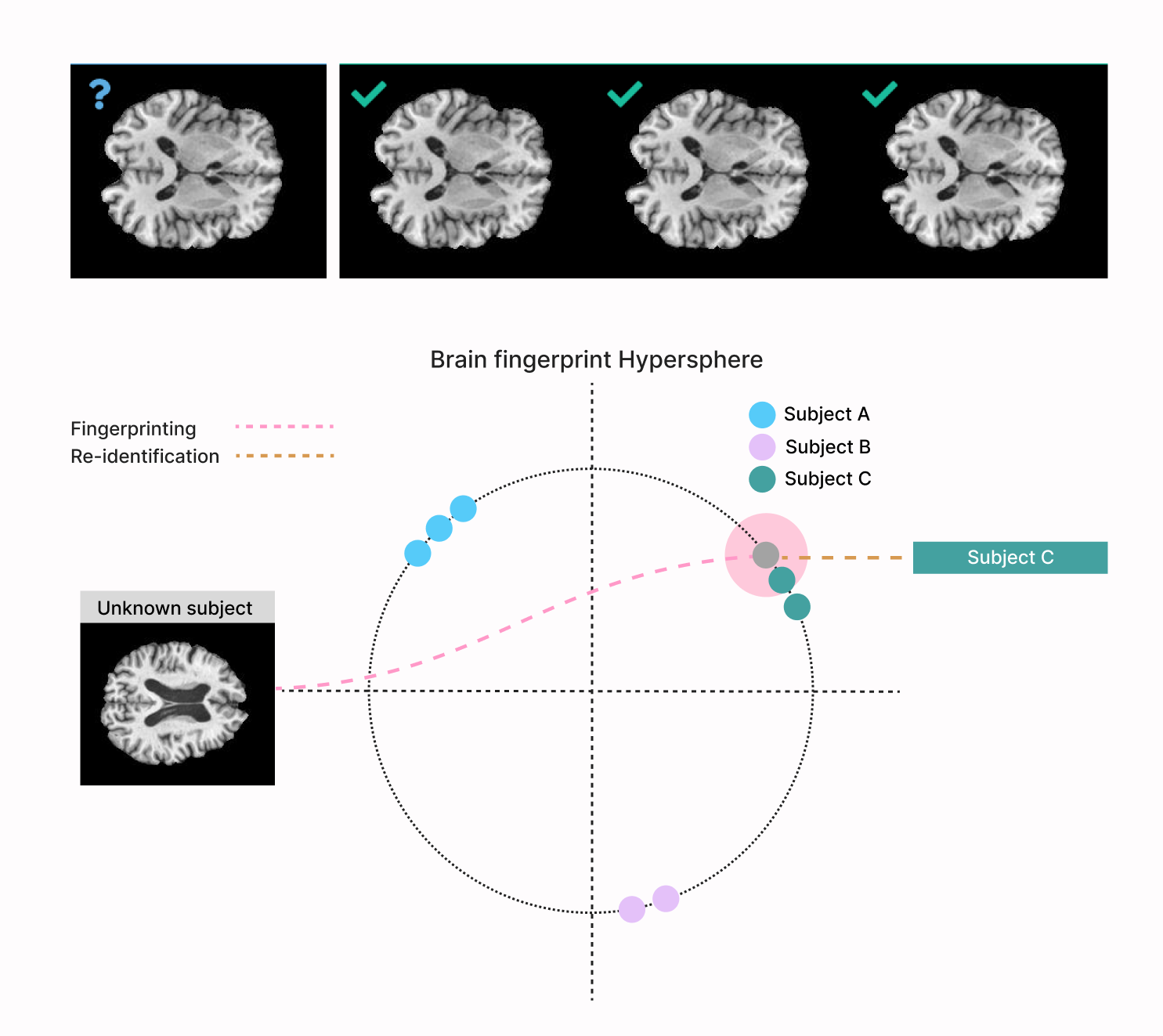 DeepBrainPrint: A Novel Contrastive Framework for Brain MRI Re-IdentificationLemuel Puglisi, Arman Eshaghi, Geoff Parker, and 3 more authorsIn Medical Imaging with Deep Learning, 2023
DeepBrainPrint: A Novel Contrastive Framework for Brain MRI Re-IdentificationLemuel Puglisi, Arman Eshaghi, Geoff Parker, and 3 more authorsIn Medical Imaging with Deep Learning, 2023Recent advances in MRI have led to the creation of large datasets. With the increase in data volume, it has become difficult to locate previous scans of the same patient within these datasets (a process known as re-identification). To address this issue, we propose an AI-powered medical imaging retrieval framework called DeepBrainPrint, which is designed to retrieve brain MRI scans of the same patient. Our framework is a semi-self-supervised contrastive deep learning approach with three main innovations. First, we use a combination of self-supervised and supervised paradigms to create an effective brain fingerprint from MRI scans that can be used for real-time image retrieval. Second, we use a special weighting function to guide the training and improve model convergence. Third, we introduce new imaging transformations to improve retrieval robustness in the presence of intensity variations (i.e. different scan contrasts), and to account for age and disease progression in patients. We tested DeepBrainPrint on a large dataset of T1-weighted brain MRIs from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and on a synthetic dataset designed to evaluate retrieval performance with different image modalities. Our results show that DeepBrainPrint outperforms previous methods, including simple similarity metrics and more advanced contrastive deep learning frameworks.